Phospho-POLR2A (S1619) Antibody
-
中文名稱:磷酸化-POLR2A (S1619)兔多克隆抗體
-
貨號:CSB-PA030114
-
規(guī)格:¥880
-
圖片:
-
其他:
產(chǎn)品詳情
-
Uniprot No.:
-
基因名:POLR2A
-
別名:DNA directed RNA polymerase II A antibody; DNA-directed RNA polymerase II largest subunit RNA polymerase II 220 kd subunit antibody; DNA-directed RNA polymerase II subunit A antibody; DNA-directed RNA polymerase II subunit RPB1 antibody; DNA-directed RNA polymerase III largest subunit antibody; hRPB220 antibody; hsRPB1 antibody; POLR2 antibody; Polr2a antibody; POLRA antibody; Polymerase (RNA) II (DNA directed) polypeptide A 220kDa antibody; Polymerase (RNA) II (DNA directed) polypeptide A antibody; RNA polymerase II subunit B1 antibody; RNA-directed RNA polymerase II subunit RPB1 antibody; RPB1 antibody; RPB1_HUMAN antibody; RPBh1 antibody; RpIILS antibody; RPO2 antibody; RPOL2 antibody
-
宿主:Rabbit
-
反應(yīng)種屬:Human,Mouse,Rat,Monkey
-
免疫原:Synthesized peptide derived from Human Rpb1 around the phosphorylation site of S1619.
-
免疫原種屬:Homo sapiens (Human)
-
標(biāo)記方式:Non-conjugated
-
抗體亞型:IgG
-
純化方式:The antibody was affinity-purified from rabbit antiserum by affinity-chromatography using epitope-specific immunogen.
-
濃度:It differs from different batches. Please contact us to confirm it.
-
保存緩沖液:Liquid in PBS containing 50% glycerol, 0.5% BSA and 0.02% sodium azide.
-
產(chǎn)品提供形式:Liquid
-
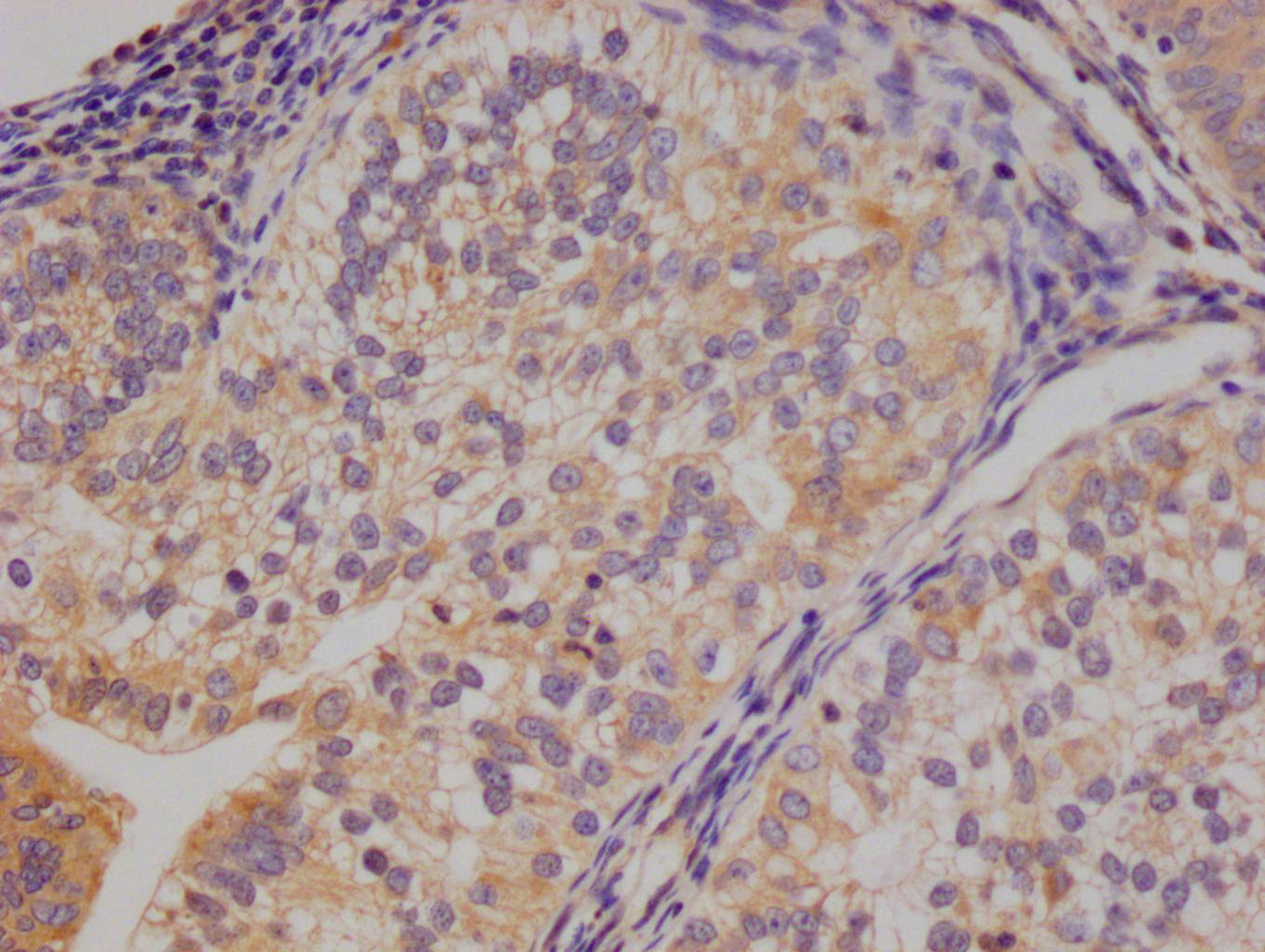
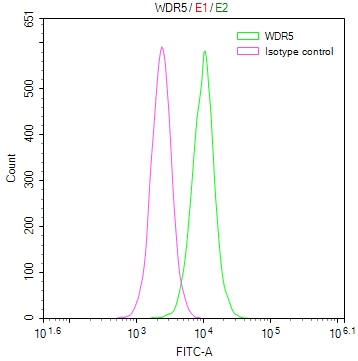
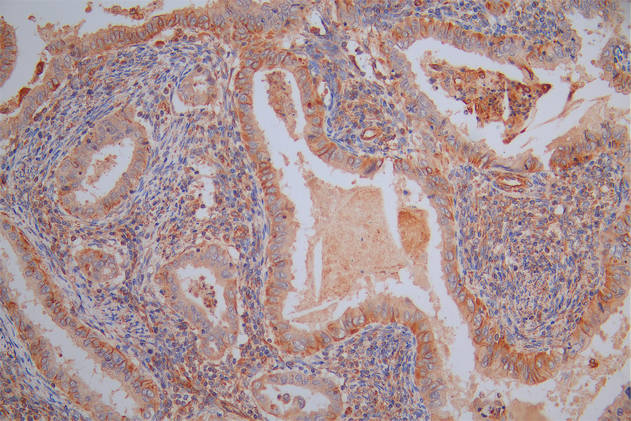
應(yīng)用范圍:WB, IHC, IF, ELISA
-
推薦稀釋比:
Application Recommended Dilution WB 1:500-1:2000 IHC 1:100-1:300 IF 1:200-1:1000 ELISA 1:10000 -
Protocols:
-
儲存條件:Upon receipt, store at -20°C or -80°C. Avoid repeated freeze.
-
貨期:Basically, we can dispatch the products out in 1-3 working days after receiving your orders. Delivery time maybe differs from different purchasing way or location, please kindly consult your local distributors for specific delivery time.
相關(guān)產(chǎn)品
靶點(diǎn)詳情
-
功能:DNA-dependent RNA polymerase catalyzes the transcription of DNA into RNA using the four ribonucleoside triphosphates as substrates. Largest and catalytic component of RNA polymerase II which synthesizes mRNA precursors and many functional non-coding RNAs. Forms the polymerase active center together with the second largest subunit. Pol II is the central component of the basal RNA polymerase II transcription machinery. It is composed of mobile elements that move relative to each other. RPB1 is part of the core element with the central large cleft, the clamp element that moves to open and close the cleft and the jaws that are thought to grab the incoming DNA template. At the start of transcription, a single-stranded DNA template strand of the promoter is positioned within the central active site cleft of Pol II. A bridging helix emanates from RPB1 and crosses the cleft near the catalytic site and is thought to promote translocation of Pol II by acting as a ratchet that moves the RNA-DNA hybrid through the active site by switching from straight to bent conformations at each step of nucleotide addition. During transcription elongation, Pol II moves on the template as the transcript elongates. Elongation is influenced by the phosphorylation status of the C-terminal domain (CTD) of Pol II largest subunit (RPB1), which serves as a platform for assembly of factors that regulate transcription initiation, elongation, termination and mRNA processing. Regulation of gene expression levels depends on the balance between methylation and acetylation levels of tha CTD-lysines. Initiation or early elongation steps of transcription of growth-factors-induced immediate early genes are regulated by the acetylation status of the CTD. Methylation and dimethylation have a repressive effect on target genes expression.; (Microbial infection) Acts as an RNA-dependent RNA polymerase when associated with small delta antigen of Hepatitis delta virus, acting both as a replicate and transcriptase for the viral RNA circular genome.
-
基因功能參考文獻(xiàn):
- XPC is an RNA polymerase II cofactor recruiting ATAC coactivator complex to promoters by interacting with E2F1. PMID: 29973595
- weak, multivalent interactions between TAF15 fibrils and heptads throughout RNA pol II CTD collectively mediate complex formation. PMID: 28945358
- This shows that CDK9 stimulates release of paused polymerase and activates transcription by increasing the number of transcribing polymerases and thus the amount of mRNA synthesized per time. PMID: 28994650
- Results identified rs2071504 in POLR2A gene to be associated with poor overall and disease-free survival of patients with an early-stage non-small cell lung cancer. PMID: 28922562
- Dara indicate that hydrogen peroxide alters RNA polymerase II (Pol II) occupancy at promoters and enhancers genome-wide. PMID: 28977633
- Rpb1/2 dynamics help govern the decision between sense and divergent antisense transcription. PMID: 28506463
- The results showed heterogeneity in the responses of individual KSHV episomes to stimuli within a single reactivating cell; those episomes that did respond to stimulation, aggregated within large domains that appear to function as viral transcription factories. A significant portion of cellular RNA polymerase II was trapped in these factories and served to transcribe viral genomes. PMID: 28331082
- Data show that inhibition of VCP/p97, or siRNA-mediated ablation of VCP/p97 impairs ultraviolet radiation (UVR)-induced RNA polymerase II (RNAPII) degradation. PMID: 28036256
- Role of chromatin-bound EGFR and ERK kinases in RNA polymerase 2 transcription PMID: 27587583
- recurrent somatic mutations in POLR2A hijack this essential enzyme and drive meningioma neoplasia PMID: 27548314
- the Elongin A ubiquitin ligase and the CSB protein function together in a common pathway in response to Pol II stalling and DNA damage PMID: 28292928
- By studying global gene expression patterns and genome-wide DNA-binding patterns of CGGBP1, it has been shown that a possible mechanism through which it affects the expression of RNA Pol II-transcribed genes in trans depends on Alu RNA. PMID: 25483050
- Using a 7,781-sample pan-cancer dataset, we first confirmed this in POLR2A are known to confer elevated sensitivity to pharmacological suppression.hese include the POLR2A interacting protein INTS10 as well as genes involved in mRNA splicing, nonsense-mediated mRNA PMID: 28027311
- HIV Tat precisely controls RNA polymerase II recruitment and pause release to fine-tune the initiation and elongation steps in target genes. PMID: 26488441
- TOP1 bound at promoters was discovered to become fully active only after pause-release. This transition coupled the phosphorylation of the carboxyl-terminal-domain (CTD) of RNA polymerase II (RNAPII) with stimulation of TOP1 above its basal rate, enhancing its processivity. PMID: 27058666
- Its variant is not related to sporadic PD in Chinese Han population. PMID: 26432391
- Data suggest RNA polymerase II (POLR2A) is extensively modified on its unique C-terminal domain (CTD) by O-GlcNAc transferase (OGT); efficient O-GlcNAcylation requires a minimum of 20 heptad CTD repeats in POLR2A and more than half of NTD of OGT. PMID: 26807597
- Serine phosphorylation stimulates whereas tyrosine phosphorylation inhibits the protein-binding activity of the RNA Pol II C-terminal domain. PMID: 26515650
- The amount of RNA polymerase II (RNAPII) on the HIV promoter and other viral regions was strongly diminished in HIV-infected CD4+ cells co-cultivated with cell non-cytotoxic antiviral response-expressing CD8+ cells. PMID: 26499373
- Ash2L acts in concert with P53 promoter occupancy to activate RNA Polymerase II by aiding formation of a stable transcription pre-initiation complex required for its activation. PMID: 25023704
- Data suggest that RNA polymerase II inhibitors may be a useful class of agent for targeting dormant leukaemia cells. PMID: 23767415
- This viral pre-initiation complex is composed of five different proteins in addition to Epstein-Barr virus BcRF1 and interacts with cellular RNA polymerase II PMID: 25165108
- Data show that E2F-1 form a complex with RNA polymerase II and protein PURA for transcriptional activation of the secondary promoter. PMID: 24819879
- human CD68 gene expression is associated with changes in Pol II phosphorylation and short-range intrachromosomal gene looping PMID: 17583472
- Authors show that the NSs protein of Schmallenberg virus (SBV) induces the degradation of the RPB1 subunit of RNA polymerase II and consequently inhibits global cellular protein synthesis and the antiviral response. PMID: 24828331
- This study reveals that TCERG1 regulates HIV-1 transcriptional elongation by increasing the elongation rate of RNAPII and phosphorylation of Ser 2 within the carboxyl-terminal domain. PMID: 24165037
- Slow Pol II elongation allows weak splice sites to be recognized, leading to higher inclusion of alternative exons. PMID: 24793692
- sequence-specific double strand DNA breaks are sufficient to activate the positive transcription elongation factor b (P-TEFb), to trigger hyperphosphorylation of the largest RNA polymerase II carboxyl-terminal-domain (Rpb1-CTD) and to induce activation of p53-transcriptional axis resulting in cell cycle arrest. PMID: 23906511
- interaction with nuclear CD26 and POLR2A gene PMID: 23638030
- RECQL5 contacts the Rpb1 jaw domain of Pol II at a site that overlaps with the binding site for the transcription elongation factor TFIIS. Binding of RECQL5 to Pol II interferes with the ability of TFIIS to promote transcriptional read-through in vitro. PMID: 23748380
- Data show that p68/DdX5 immunoprecipitated with RNA polymerase II (RNAP II) and suggest p68 is important in facilitating beta-catenin and androgen receptor (AR) transcriptional activity in prostate cancer cells. PMID: 23349811
- inhibition of the transition of paused RNA PolII to productive elongation, described here for p21(CIP1), is a general mechanism by which transcription factor Sp3 fine-tunes gene expression. PMID: 23401853
- RNA polymerase II acts as an RNA-dependent RNA polymerase to extend and destabilize a non-coding RNA. PMID: 23395899
- Data indicate that polyamide treatment activates p53 signaling and results in a time- and and dose-dependent depletion of the RNA polymerase II (RNAP2) large subunit RPB1. PMID: 23319609
- CTCF binding sites regulate mRNA production, RNA polymerase II (RNAPII) programming, and nucleosome organization of the Kaposi's sarcoma-associated herpesvirus latency transcript control region. PMID: 23192870
- site-specific p65 phosphorylation targets NF-kappaB activity to particular gene subsets on a global level by influencing p65 and p-RNAP II promoter recruitment PMID: 23100252
- BRD4-driven Pol II phosphorylation at serine 2 plays an important role in regulating lineage-specific gene transcription in human CD4+ T cells. PMID: 23086925
- SNAPC1 is a general transcriptional coactivator that functions through elongating RNAPII. PMID: 22966203
- Cyclin K1 is the primary cyclin partner for CDK12/CrkRS and it is required for activation of CDK12/CrkRS to phosphorylate the C-terminal domain of RNA Pol II. PMID: 22988298
- Studies indicate that the super elongation complex (SEC) consisting of ELL, P-TEFb (CDK9) and MLL required for rapid transcriptional induction in the presence or absence of paused RNA polymerase II (Pol II). PMID: 22895430
- Results indicate roles for both the RNA polymerase II C-terminal domain (CTD) and O-GlcNAc in the regulation of transcription initiation. PMID: 22605332
- Here, the authors report phosphorylation of Thr4 by Polo-like kinase 3 in mammalian cells. PMID: 22549466
- Studies suggest activator-induced structural shifts within Mediator trigger activation of stalled Pol II. PMID: 21326907
- These results suggest that Mediator structural shifts induced by activator binding help stably orient pol II prior to transcription initiation within the human mediator-RNA polymerase II-TFIIF assembly. PMID: 22343046
- evidence that phosphorylation of Rpb1 CTD Thr4 residues is required specifically for histone mRNA 3' end processing, functioning to facilitate recruitment of 3' processing factors to histone genes PMID: 22053051
- Parcs/Gpn3 plays a critical role in the nuclear accumulation of RNAP II, and this function explains the relative importance of Parcs/Gpn3 in cell proliferation. PMID: 21782856
- kinetics of RNA polymerase II elongation during co-transcriptional splicing PMID: 21264352
- Data show that MicroRNA promoter identification based upon RPol II binding patterns provides important temporal and spatial measurements regarding the initiation of transcription. PMID: 21072189
- The deregulation of cellular NIPP1/PP1 holoenzyme affects RNAPII phosphorylation and pointing to NIPP1 as a potential regulatory factor in RNAPII-mediated transcription. PMID: 20941529
- Elevated PHD1 concomitant with decreased PHD2 are causatively related to Rpb1 hydroxylation and oncogenesis in human renal clear cell carcinomas with WT VHL gene. PMID: 20978146
顯示更多
收起更多
-
亞細(xì)胞定位:Nucleus. Cytoplasm. Chromosome.
-
蛋白家族:RNA polymerase beta' chain family
-
數(shù)據(jù)庫鏈接: