DDX5 Recombinant Monoclonal Antibody
-
中文名稱:DDX5重組抗體
-
貨號:CSB-RA231179A0HU
-
規(guī)格:¥1320
-
圖片:
-
Western Blot
Positive WB detected in: HepG2 whole cell lysate, HT-29 whole cell lysate, 293T whole cell lysate, Jurkat whole cell lysate, Mouse brain tissue
All lanes: DDX5 antibody at 1:2000
Secondary
Goat polyclonal to rabbit IgG at 1/50000 dilution
Predicted band size: 70, 61 kDa
Observed band size: 70 kDa -
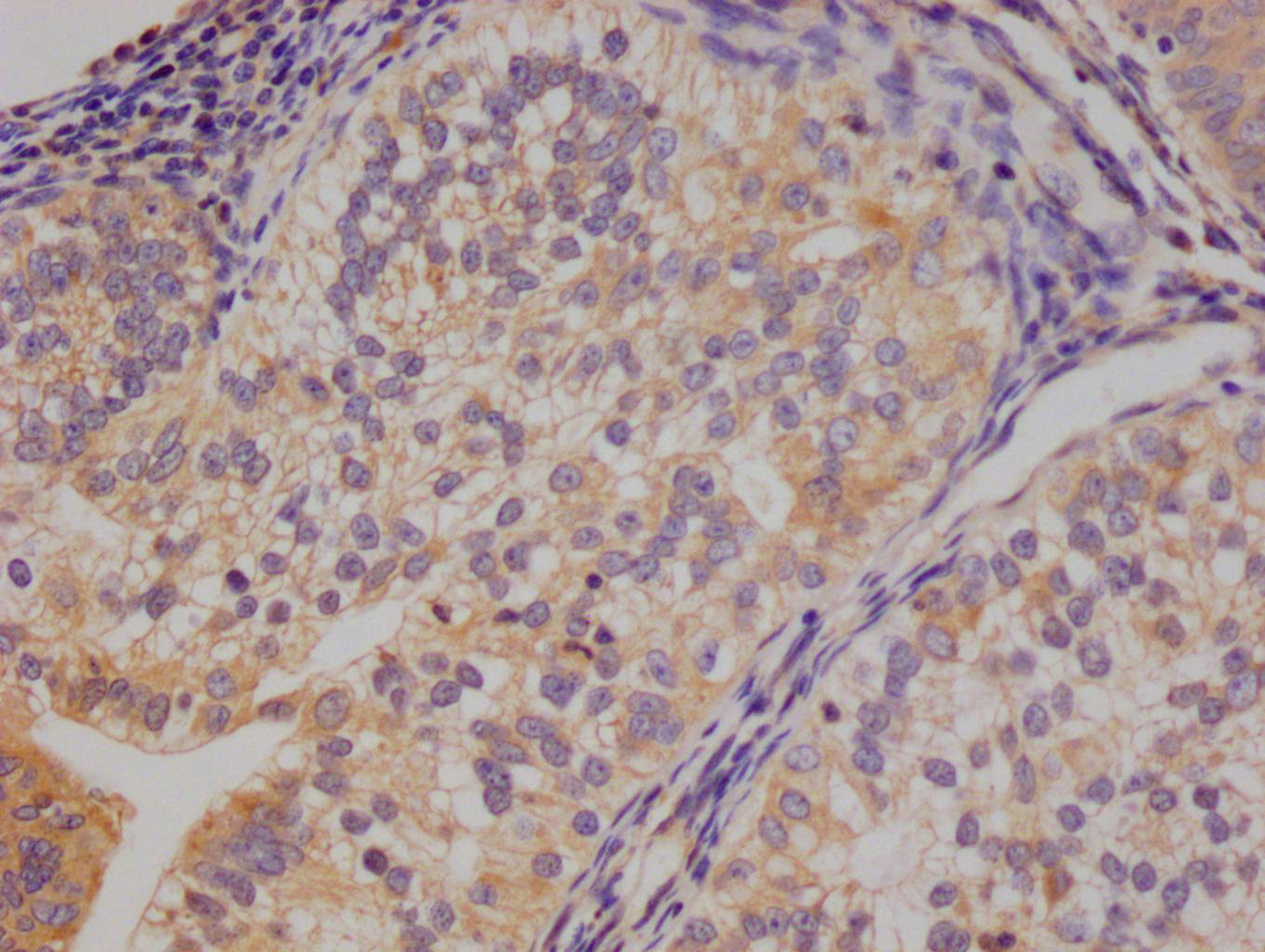
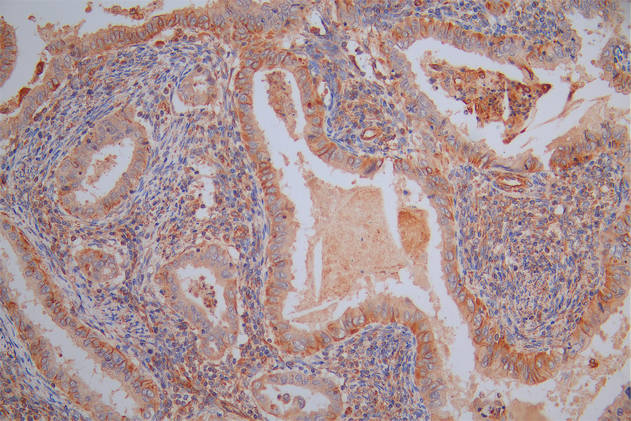
IHC image of CSB-RA231179A0HU diluted at 1:100 and staining in paraffin-embedded human colon cancer performed on a Leica BondTM system. After dewaxing and hydration, antigen retrieval was mediated by high pressure in a citrate buffer (pH 6.0). Section was blocked with 10% normal goat serum 30min at RT. Then primary antibody (1% BSA) was incubated at 4℃ overnight. The primary is detected by a Goat anti-rabbit IgG polymer labeled by HRP and visualized using 0.05% DAB.
-
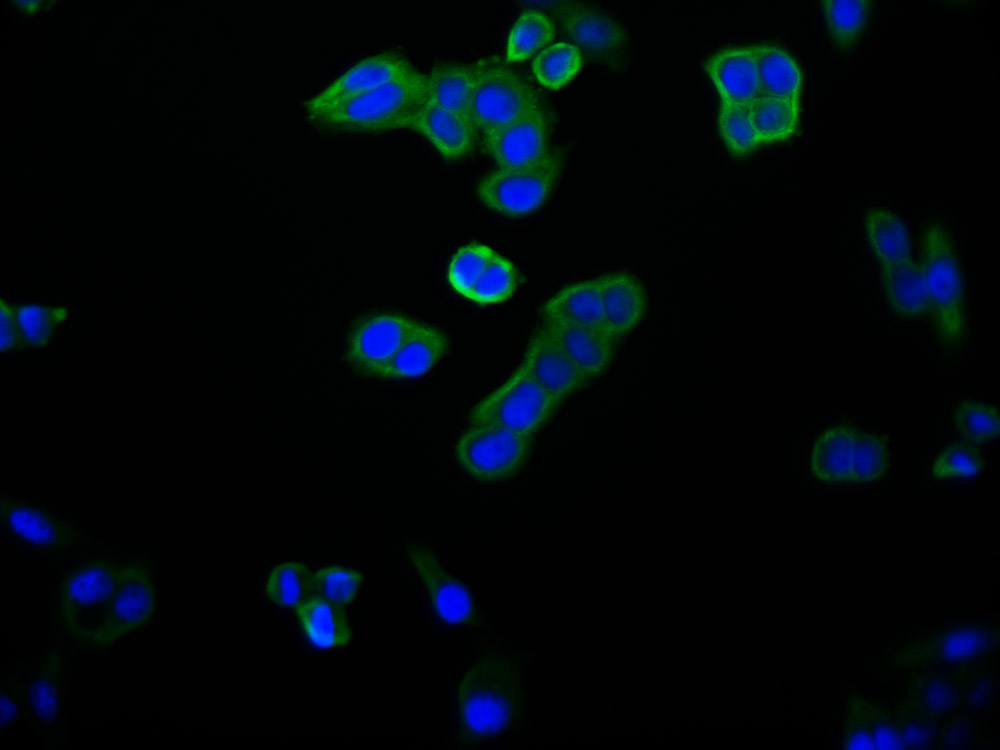
Immunofluorescence staining of HepG2 Cells with CSB-RA231179A0HU at 1:50, counter-stained with DAPI. The cells were fixed in 4% formaldehyde, permeated by 0.2% TritonX-100, and blocked in 10% normal Goat Serum. The cells were then incubated with the antibody overnight at 4℃. Nuclear DNA was labeled in blue with DAPI. The secondary antibody was FITC-conjugated AffiniPure Goat Anti-Rabbit IgG (H+L).
-
Overlay histogram showing Hela cells stained with CSB-RA231179A0HU (red line) at 1:50. The cells were fixed with 70% Ethylalcohol (18h) and then incubated in 10% normal goat serum to block non-specific protein-protein interactions followedby the antibody (1μg/1*106 cells) for 1 h at 4℃.The secondary antibody used was FITC-conjugated goat anti-rabbit IgG (H+L) at 1/200 dilution for 30min at 4℃. Control antibody (green line) was Rabbit IgG (1μg/1*106 cells) used under the same conditions. Acquisition of >10,000 events was performed.
-
-
其他:
產(chǎn)品詳情
-
產(chǎn)品描述:
DDX5 is an ATP-dependent RNA helicase involved in splicing, mRNA export, transcript stability, rRNA biosynthesis, and microRNA processing, among other RNA-related events. DDX5 has been connected to the regulation of numerous cancer-related pathways and has been found to be abnormally expressed in many cancers. DDX5 is closely linked to carcinogenesis, invasiveness, and metastasis, as well as cancer proliferation in several cancers. Dysfunction of DDX5 is ultimately associated with tumor formation and progression.
CUSABIO designed the vector clones for the expression of a recombinant DDX5 antibody in mammalian cells. The vector clones were obtained by inserting the DDX5 antibody heavy and light chains into the plasma vectors. The recombinant DDX5 antibody was purified from the culture medium through Affinity-chromatography. It can be used to detect DDX5 protein from Human, Mouse in the ELISA, WB, IHC, IF, FC. -
Uniprot No.:
-
基因名:
-
別名:Probable ATP-dependent RNA helicase DDX5 (EC 3.6.4.13) (DEAD box protein 5) (RNA helicase p68), DDX5, G17P1 HELR HLR1
-
反應(yīng)種屬:Human, Mouse
-
免疫原:A synthesized peptide derived from human DDX5
-
免疫原種屬:Homo sapiens (Human)
-
標(biāo)記方式:Non-conjugated
-
克隆類型:Monoclonal
-
抗體亞型:Rabbit IgG
-
純化方式:Affinity-chromatography
-
克隆號:2C3
-
濃度:It differs from different batches. Please contact us to confirm it.
-
保存緩沖液:Rabbit IgG in phosphate buffered saline, pH 7.4, 150mM NaCl, 0.02% sodium azide and 50% glycerol.
-
產(chǎn)品提供形式:Liquid
-
應(yīng)用范圍:ELISA, WB, IHC, IF, FC
-
推薦稀釋比:
Application Recommended Dilution WB 1:500-1:5000 IHC 1:50-1:200 IF 1:20-1:200 FC 1:20-1:200 -
Protocols:
-
儲存條件:Upon receipt, store at -20°C or -80°C. Avoid repeated freeze.
-
貨期:Basically, we can dispatch the products out in 1-3 working days after receiving your orders. Delivery time maybe differs from different purchasing way or location, please kindly consult your local distributors for specific delivery time.
相關(guān)產(chǎn)品
靶點(diǎn)詳情
-
功能:Involved in the alternative regulation of pre-mRNA splicing; its RNA helicase activity is necessary for increasing tau exon 10 inclusion and occurs in a RBM4-dependent manner. Binds to the tau pre-mRNA in the stem-loop region downstream of exon 10. The rate of ATP hydrolysis is highly stimulated by single-stranded RNA. Involved in transcriptional regulation; the function is independent of the RNA helicase activity. Transcriptional coactivator for androgen receptor AR but probably not ESR1. Synergizes with DDX17 and SRA1 RNA to activate MYOD1 transcriptional activity and involved in skeletal muscle differentiation. Transcriptional coactivator for p53/TP53 and involved in p53/TP53 transcriptional response to DNA damage and p53/TP53-dependent apoptosis. Transcriptional coactivator for RUNX2 and involved in regulation of osteoblast differentiation. Acts as transcriptional repressor in a promoter-specific manner; the function probably involves association with histone deacetylases, such as HDAC1. As component of a large PER complex is involved in the inhibition of 3' transcriptional termination of circadian target genes such as PER1 and NR1D1 and the control of the circadian rhythms.
-
基因功能參考文獻(xiàn):
- RIP-seq analysis in HEK293T cells identifies a complete repertoire of DDX5/p68 interacting transcripts including LOC284454 lncRNA. PMID: 29227193
- Results showed that DDX5 was significantly up-regulated in gastric cancer tissues and revealed a novel role of DDX5 in gastric cancer cell proliferation via the mTOR pathway. PMID: 28216662
- DDX5 is shown to be involved in RNA metabolism and viral infection, especially for RNA virus which seems to hijack host DDX5 to facilitate its own replication. However, DDX5 exhibits a role in antiviral responses during HBV and MYXV infection. Its opposite roles between DNA and RNA infection likely reflect the different modes of the biosynthesis of RNA and DNA viruses. [review] PMID: 29642538
- A significant overlap between hnRNPA1 and DDX5 splicing targets and they share many closely linked binding sites. PMID: 30042133
- The role of DDX5 in regulating esophageal cancer cell proliferation and tumorigenesis.DDX5 is highly expressed in esophageal cancer. PMID: 28244855
- Downregulation of p68 RNA Helicase (DDX5) Activates a Survival Pathway Involving mTOR and MDM2 Signals.( PMID: 28557706
- Results show refined biochemical and biological comparison of yeast Dbp2 and human DDX5 enzymes. Human DDX5 possesses a 10-fold higher unwinding activity than Dbp2, partially due to the presence of a mammalian/avian specific C-terminal extension. Also, ectopic expression of DDX5 rescues the cold sensitivity, cryptic initiation defects, and impaired glucose import in dbp2Delta cells, suggesting functional conservation. PMID: 28411202
- p53 gain-of-function mutations accelerate endometrial carcinoma progression and metastasis by interfering with Drosha and p68 binding and pri-miR-26a-1 processing, resulting in reduced miR-26a expression and EZH2 overexpression. PMID: 26587974
- Cervical cancer cell DDX5 gene is transcriptionally upregulated by calcitriol through a VDRE located in its proximal promoter. PMID: 26314252
- Systematic Determination of Human Cyclin Dependent Kinase (CDK)-9 Interactome Identifies Novel Functions in RNA Splicing Mediated by the DEAD Box DDX5 and DDX17 RNA Helicases PMID: 26209609
- LMTK3 escapes tumour suppressor miRNAs via sequestration of DDX5. PMID: 26739063
- The data provide a model in which p68 and p53 interplay regulates PLK1 expression, and which describes the behavior of these molecules, and the outcome of their interaction, in human breast cancer. PMID: 24626184
- Results show that a new mechanism of oncogenesis is attributed to p68 by upregulation of AKT and consequent nuclear exclusion and degradation of tumor suppressor FOXO3a. PMID: 25745998
- DDX5 played an important role in the proliferation and tumorigenesis of non-small-cell lung cancer cells by activating the beta-catenin signaling pathway. PMID: 26212035
- study shows that correction of p68 may reduce toxicity of the mutant RNAs in DM1 and in DM2 PMID: 26080402
- Data indicate that cyclooxygenase 2 (COX-2) correlates inversely to microRNA 183 (miR-183) and directly to DEAD-box helicase p68 (DDX5). PMID: 25963660
- DDX5 protein is essential for normal cell proliferation; (2) the transition from G1 to S/G2 phase is accompanied by an increase of DDX5 protein concentration in the cells. PMID: 26035968
- AML is dependent on DDX5 and that inhibiting DDX5 expression slows AML cell proliferation. PMID: 24910429
- Downregulation of DDX5 and DDX17 protein expression during myogenesis and epithelial-to-mesenchymal transdifferentiation contributes to the switching of splicing programs during these processes. PMID: 24910439
- Data indicate that armadillo repeat protein ARVCF interacts with the splicing factors the splicing factor SRSF1 (SF2/ASF), the RNA helicase p68 (DDX5), and the heterogeneous nuclear ribonucleoprotein hnRNP H2. PMID: 24644279
- In conclusion, authors identified DDX5 as a positive regulator for Japanese encephalitis virus replication via its binding to viral 3' UTR. PMID: 24035833
- DDX5 facilitates HIV-1 replication as a cellular co-factor of Rev. PMID: 23741449
- DDX5 might be critical for NOTCH1-mediated T-ALL pathogenesis and thus is a potential new target for modulating the Notch signaling in leukemia PMID: 23108395
- p68, in the presence of Ca-calmodulin, can function as a microtubule motor. PMID: 23322042
- Results show a novel role for DDX5 in cancer cell proliferation and suggest DDX5 as a therapeutic target in breast cancer treatment. PMID: 22750847
- High p68 RNA helicase expression is associated with glioma. PMID: 22810421
- Data indicate that transcriptional coregulator ddx5/ddx17 RNA helicases can simultaneously regulate the transcriptional activity and alternative splicing of NFAT5 transcription factor. PMID: 22266867
- RNA helicases Ddx17 and Ddx5 contribute to tumor-cell invasiveness by regulating alternative splicing of several DNA- and chromatin-binding factors, including the macroH2A1 histone. PMID: 23022728
- The DEAD box RNA helicase p68, also referred to as DDX5, directly interacts with VDR. PMID: 22476084
- there is a direct interaction between DDX5 and NS5B and DDX5 has two independent NS5B-binding sites PMID: 22640416
- High DDX5 is associated with basal breast cancer cells. PMID: 22086602
- Using an RNA affinity pulldown-coupled mass spectrometry approach the study identified DDX5/RNA helicase p68 as an activator of TAU exon 10 splicing. PMID: 21343338
- a striking inverse association of p68 and delta133p53 expression in primary breast cancers was identified. PMID: 20818423
- DEAD-box RNA helicase p68 (DDX5) and its associated noncoding RNA, steroid receptor RNA activator (SRA), form a complex with CTCF that is essential for insulator function PMID: 20966046
- Pleiotropic effects of p300-mediated acetylation on p68 and p72 RNA helicase. PMID: 20663877
- crystallization and preliminary diffraction analysis of N-terminal region of DDX5 is reported.X-ray diffraction data were processed to a resolution of 2.7 A. PMID: 20124720
- DDX5 is a repressor of fibrogenic genes in HSCs through interaction with transcriptional complexes. PMID: 20022962
- Essential for pre-mrna splicing in vitro; may function in destabilizing the U1-5'ss interaction. Depletion of p68 RNA helicase arrested spliceosome assembly at the prespliceosome stage PMID: 12101238
- synergism with transcriptional coactivators CBP and p300 PMID: 12527917
- role in c-H-ras alternative splicing regulation PMID: 12665590
- p68 is an important transcriptional regulator, functioning as a co-activator and/or co-repressor depending on the context of the promoter & the transcriptional complex. AA 1-478 of p68 can repress transcription as well as the full-length protein. PMID: 15298701
- there is a tightly controlled expression and nucleolar localization of p68 in keratinocytes in vitro and during skin repair in vivo that functionally contributes to keratinocyte proliferation and gene expression PMID: 15304501
- mechanism by which p68 may act as a tumour cosuppressor in governing p53 transcriptional activity PMID: 15660129
- data suggest that function(s) of p68 RNA helicase may be subjected to the regulation of multiple cell signal pathways PMID: 15927448
- In addition, it could be demonstrated that increasing the Tlk1 activity in HT1080 cells by forced Tlk1 overexpression leads to an increased phosphorylation of endogenous p68. PMID: 15950181
- Patient with chronic hepatitis C carrying DDX5 haplotypes are at an increased risk of developing advanced liver fibrosis. PMID: 16697732
- SUMO modification of the DEAD box protein p68 modulates its transcriptional activity and promotes its interaction with HDAC1 PMID: 17369852
- A mutant that carries mutations at the phosphorylation sites (Y593/595F) dramatically sensitizes TRAIL-resistant cells to TRAIL-induced apoptosis, suggesting a potential therapeutic strategy to overcome TRAIL resistance. PMID: 17384675
- The percentage correlation between Q-RT-PCR and microarray were 70% and 48% by using DDX5 and GAPDH as internal controls, respectively. PMID: 17540040
- p68/p72 may contribute to colon cancer formation by directly up-regulating proto-oncogenes and indirectly by down-regulating the growth suppressor p21(WAF1/CIP1). PMID: 17699760
顯示更多
收起更多
-
亞細(xì)胞定位:Nucleus. Nucleus, nucleolus. Cytoplasm.
-
蛋白家族:DEAD box helicase family, DDX5/DBP2 subfamily
-
數(shù)據(jù)庫鏈接:
Most popular with customers
-
-
YWHAB Recombinant Monoclonal Antibody
Applications: ELISA, WB, IF, FC
Species Reactivity: Human, Mouse, Rat
-
Phospho-YAP1 (S127) Recombinant Monoclonal Antibody
Applications: ELISA, WB, IHC
Species Reactivity: Human
-
-
-
-
-