Recombinant Saccharomyces cerevisiae Exodeoxyribonuclease 1 (EXO1), partial
-
中文名稱:釀酒酵母EXO1重組蛋白
-
貨號(hào):CSB-YP331092SVG
-
規(guī)格:
-
來源:Yeast
-
其他:
-
中文名稱:釀酒酵母EXO1重組蛋白
-
貨號(hào):CSB-EP331092SVG
-
規(guī)格:
-
來源:E.coli
-
其他:
-
中文名稱:釀酒酵母EXO1重組蛋白
-
貨號(hào):CSB-EP331092SVG-B
-
規(guī)格:
-
來源:E.coli
-
共軛:Avi-tag Biotinylated
E. coli biotin ligase (BirA) is highly specific in covalently attaching biotin to the 15 amino acid AviTag peptide. This recombinant protein was biotinylated in vivo by AviTag-BirA technology, which method is BriA catalyzes amide linkage between the biotin and the specific lysine of the AviTag.
-
其他:
-
中文名稱:釀酒酵母EXO1重組蛋白
-
貨號(hào):CSB-BP331092SVG
-
規(guī)格:
-
來源:Baculovirus
-
其他:
-
中文名稱:釀酒酵母EXO1重組蛋白
-
貨號(hào):CSB-MP331092SVG
-
規(guī)格:
-
來源:Mammalian cell
-
其他:
產(chǎn)品詳情
-
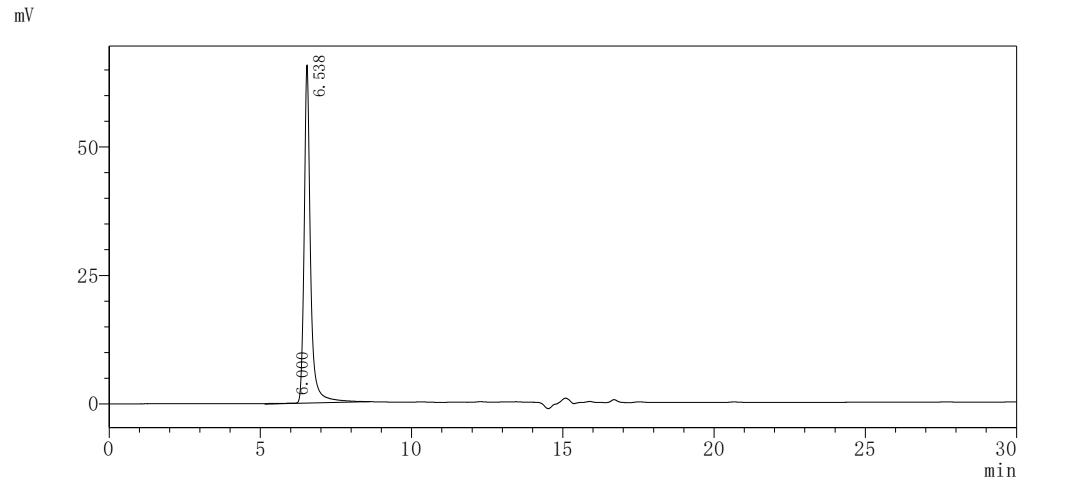
純度:>85% (SDS-PAGE)
-
基因名:
-
Uniprot No.:
-
別名:EXO1; DHS1; YOR033C; OR26.23Exodeoxyribonuclease 1; EC 3.1.-.-; Exodeoxyribonuclease I; EXO I; Exonuclease I; Protein DHS1
-
種屬:Saccharomyces cerevisiae (strain ATCC 204508 / S288c) (Baker's yeast)
-
蛋白長(zhǎng)度:Partial
-
蛋白標(biāo)簽:Tag?type?will?be?determined?during?the?manufacturing?process.
The tag type will be determined during production process. If you have specified tag type, please tell us and we will develop the specified tag preferentially. -
產(chǎn)品提供形式:Lyophilized powder
Note: We will preferentially ship the format that we have in stock, however, if you have any special requirement for the format, please remark your requirement when placing the order, we will prepare according to your demand. -
復(fù)溶:We recommend that this vial be briefly centrifuged prior to opening to bring the contents to the bottom. Please reconstitute protein in deionized sterile water to a concentration of 0.1-1.0 mg/mL.We recommend to add 5-50% of glycerol (final concentration) and aliquot for long-term storage at -20℃/-80℃. Our default final concentration of glycerol is 50%. Customers could use it as reference.
-
儲(chǔ)存條件:Store at -20°C/-80°C upon receipt, aliquoting is necessary for mutiple use. Avoid repeated freeze-thaw cycles.
-
保質(zhì)期:The shelf life is related to many factors, storage state, buffer ingredients, storage temperature and the stability of the protein itself.
Generally, the shelf life of liquid form is 6 months at -20°C/-80°C. The shelf life of lyophilized form is 12 months at -20°C/-80°C. -
貨期:Delivery time may differ from different purchasing way or location, please kindly consult your local distributors for specific delivery time.Note: All of our proteins are default shipped with normal blue ice packs, if you request to ship with dry ice, please communicate with us in advance and extra fees will be charged.
-
注意事項(xiàng):Repeated freezing and thawing is not recommended. Store working aliquots at 4°C for up to one week.
-
Datasheet :Please contact us to get it.
靶點(diǎn)詳情
-
功能:5'->3' double-stranded DNA exonuclease involved in mismatch repair and eventually also in mitotic recombination between direct repeats. Also has a minor role in the correction of large DNA mismatches that occur in the heteroduplex DNA during meiotic recombination at the HIS4 locus.
-
基因功能參考文獻(xiàn):
- Exo1-dependent and independent mismatch repair has been described. (Review) PMID: 25956862
- When RNR is induced in the absence of Exo1 and RNR negative regulators, cell viability of rad53 mutants treated with HU is increased and the ability of replication forks to restart after replicative stress is restored. PMID: 25601385
- Exo1 function in the DNA-damage response and the regulation of these functions by phosphorylation PMID: 25457771
- We show that the loss of Exo1p reduces the rate of UV-induced, but not spontaneous, recombination events and that the lengths of crossover-associated conversion events are reduced in the exo1 strain. PMID: 24835424
- Occurrence of duplication of the targeted chromosome was strikingly increased in the exo1Delta sgs1Delta double mutant but not in the respective single mutants. PMID: 25089886
- Models for the functional interactions of mitotic cyclin Clb2, Sgs1 and Exo1 with replication fork stabilization are proposed. PMID: 25771727
- Characterization of the sensitivity of single, double and triple mutants for Exo1, Mre11 and Pso2 nucleases in Saccharomyces cerevisiae to various DNA damaging agents reveals complex interactions that depend on the type of DNA damage. PMID: 18295552
- Deletion of Exo1 reduces efficiency of mismatch repair of errors made by Pol1 even more than for errors made by Pol3. PMID: 23245696
- A prototypical Franconi anemia-related interstrand crosslink repair pathway operates in budding yeast, which acts redundantly with the pathway controlled by Pso2, and is required for the targeting of Exo1 to chromatin to execute DNA repair. PMID: 22912599
- Data provide a model that indicates how in Rad53 or MRX mutants, an inappropriately active Exo1 may facilitate faulty template switching between nearby inverted repeats to form dicentric chromosomes. PMID: 21098663
- Mre11 and Exo1 are the major nucleases involved in creating resection tracts of widely varying lengths typical of meiotic recombination. PMID: 21146476
- Exo1 also plays a nuclease-independent role in crossover promotion. PMID: 21044871
- Exo1 processes nucleotide excision repair (NER) intermediates when repair synthesis is impeded, converting NER intermediates to long ssDNA gaps and promoting checkpoint activation. PMID: 20932474
- In the absence of end protection by Ku, the requirement for the MRX complex is bypassed and 5'-3' resection of double-strand break ends is executed by Exo1. PMID: 20729809
- Exo1 exonuclease is an initiator of the recombination process that allows cells to escape senescence and become immortal in the absence of telomerase. PMID: 15126386
- Exo1 can make either positive or negative contributions to telomere function and cell viability, depending on whether telomerase or recombination is utilized to maintain telomere function. PMID: 15126387
- Exo1 is critical for generating ssDNA in subtelomeric X repeats and internal single-copy sequences of Saccharomyces cerevisiae cdc13-1 mutants. PMID: 15454530
- Exo1 exonuclease is recruited to stalled forks and, in rad53 mutants, counteracts reversed fork accumulation PMID: 15629726
- Exo1p plays both structural and catalytic roles during DNA mismatch repair. PMID: 17602897
- SNPs associated with prognosis of lung cancer was mapped to EXO1. PMID: 17855454
- Rad53-dependent Exo1 phosphorylation is involved in a negative feedback loop to limit ssDNA accumulation and DNA damage checkpoint activation PMID: 18756267
- Deletion of SGS1 or DNA2 reduces resection and DSB repair by single-strand annealing between distant repeats while the remaining long-range resection activity depends on the exonuclease Exo1. PMID: 18805091
- yeast Exo1 nuclease and Sgs1 helicase function in alternative pathways for double strand break processing PMID: 18806779
- The site S2 in Mlh1 mediates Exo1 recruitment in order to optimize mismatch repair-dependent mutation avoidance. PMID: 19015241
- Data further show that Dna2 might support Sgs1 activity, as it acts redundantly with Exo1, but not with Sgs1 in telomere processing. PMID: 19595717
顯示更多
收起更多
-
亞細(xì)胞定位:Nucleus.
-
蛋白家族:XPG/RAD2 endonuclease family, EXO1 subfamily
-
數(shù)據(jù)庫鏈接:
KEGG: sce:YOR033C
STRING: 4932.YOR033C
Most popular with customers
-
Recombinant Human Alkaline phosphatase, germ cell type (ALPG) (Active)
Express system: Mammalian cell
Species: Homo sapiens (Human)
-
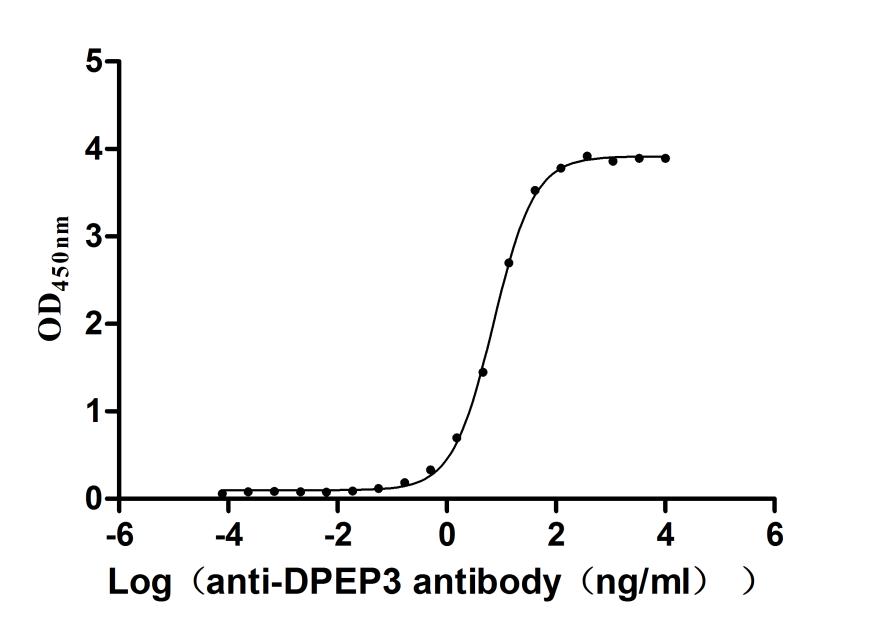
Recombinant Human Dipeptidase 3(DPEP3), partial (Active)
Express system: Mammalian cell
Species: Homo sapiens (Human)
-
Recombinant Human C-C chemokine receptor type 9 (CCR9)-VLPs (Active)
Express system: Mammalian cell
Species: Homo sapiens (Human)
-
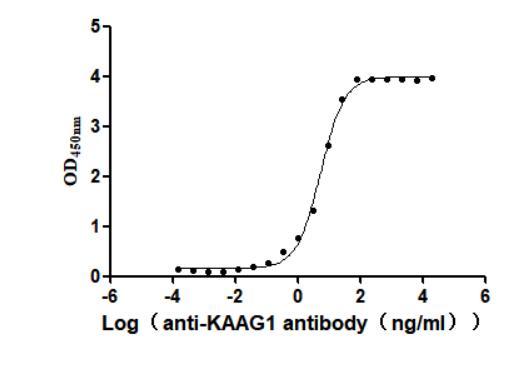
Recombinant Human Kidney-associated antigen 1(KAAG1) (Active)
Express system: Baculovirus
Species: Homo sapiens (Human)